Really grateful to get the opportunity to co-teach DSCI 303: Machine Learning for Data Science in tandem with Dr. Akane Sano. We tried to teach fundamental ML concepts and techniques from a conceptual, less math-y framework. The students worked on several exciting projects in the domains of healthcare, entertainment, and even gaming!
Category Archives: Uncategorized
Postdoc at Scalable Health Labs
I officially started a postdoctoral research scientist position at Rice’s Scalable Health labs under Dr. Ashutosh Sabharwal on August 1. I look forward to continuing my work in using machine learning, signal processing, and statistics to find clinically meaningful signatures in medical imaging and sensor data. I am especially excited to be focusing more on pediatric disorders, and looking to contribute to the nascent field of computational pediatrics!
Doc!
Spatial infiltration analysis work in Inflammatory Breast Cancer accepted in Cancer Immunology
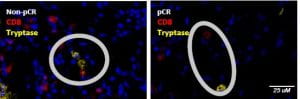
The spatial proximity of mast cells (Tryptase) and CD8+ T-cells was significantly associated with pathologic complete response (pCR) to neoadjuvant chemotherapy regimens.
Work we did with Dr. Sangeetha Reddy and Dr. Jennifer Wargo to analyze the spatial relationships amongst various cell types in IBC is published in Cancer Immunology. Inflammatory breast cancer (IBC) is an aggressive form of primary cancer with low rates of pathologic complete response to current neoadjuvant chemotherapy (NAC) regimens. Our spatial analysis framework showed the close proximity of mast cells to CD8+ T cells, CD163+ monocytes/macrophages, and tumor cells when a pathologic complete response was not achieved. The proximity of mast cells to immune and tumor cells may suggest immunosuppressive or tumor-promoting interactions of these mast cells, and these can be a potential therapeutic target in IBC. Read more here: http://cancerimmunolres.aacrjournals.org/content/7/6/1025.abstract
Image courtesy Dr. Sangeetha Reddy
Defended my Ph.D.!
It is finally done! I defended my Ph.D. thesis on March 20, 2019. Big thank you to my advisor Dr. Arvind Rao and my committee chair Dr. Ashok Veeraraghavan for their invaluable support, guidance, and mentorship during the last five and a half years. Thanks to all my friends and family, without whom this day would not have been possible. 🙂
G-function co-localization work in Merkel Cell Carcinoma gets in Clinical Cancer Research
In this paper, we propose a co-localization metric derived from the G-function and show that this metric is significantly correlated with adverse tumor signatures, such as larger tumor size, increased vascular density, and greater depth of invasion. Pre-print to be uploaded soon! This work was done in collaboration with the wonderful Dr. Michael Tetzlaff from the Department of Pathology at the M.D Anderson Cancer Center.
Switzerland gallery
- By Lake Geneva
- Swiss-Italian lunch
- From the summit at Uetliberg
- Lake Zurich backdrop
Attended inaugural PhD summit at EPFL, Switzerland
I was chosen as one of 13 attendees worldwide for the inaugural PhD summit at EPFL, Lausanne, Switzerland. The summit was designed to bring together final year PhD students to present and discuss their work with EPFL professors and students. The theme of this year was ‘Data-driven engineering in Life sciences’ (https://phdsummit.epfl.ch/)
I presented my work on ‘Towards Personalized Treatment: Leveraging structure in cancer imaging to predict clinical outcomes’. A video of my talk can be found here: https://phdsummit.epfl.ch/presentations/
Overall I had a blast interacting with exceptional students from the US, Singapore, and different parts of Europe. 
Very grateful to the EPFL PhD summit organizing committee, especially Drs. Ali Sayed for the initiative, Pascal Frossard and Jose Millan for the lab visits, and the tireless and amazing Sebastian Gautsch for managing anything and everything for this trip.
Functional Spatial Analysis for risk assessment in IPMN published in Cancer Informatics
Our paper titled “A Functional Spatial Analysis Platform for Discovery of Immunological Interactions Predictive of Low-Grade to High-Grade Transition of Pancreatic Intraductal Papillary Mucinous Neoplasms” has been published in Cancer Informatics’ Special issue on Ensemble Learning and Deep Learning in Cancer Genomic and Imaging Data. In this work, we enhance our G-function based spatial analysis framework by incorporating ideas from functional data analysis, to build a tool more effective at representing the structure in the G-function. We show that spatial metrics derived from our functional spatial platform is able to predict the risk of progression in IPMN’s with a higher accuracy than simpler metrics such as counts or the G-function AUC alone. AN ensemble of models built using counts and the proposed G-function MFPCA metric performs the best. The paper can be found here.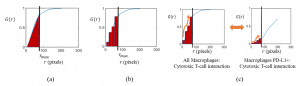
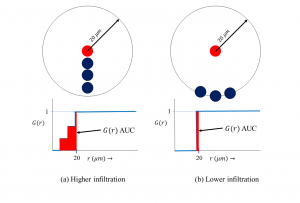
Spatial tumor:Treg interaction predicts poor survival in Non-Small Cell Lung Cancer accepted to Lung Cancer journal
Our paper on using the spatial G-function to quantify tumor:immune interactions in multiplexed Immunofluorescence images from Non-Small Cell Lung Cancer patients has been accepted to Lung Cancer! [Paper link] We made two contributions in this paper: 1) Repurposed the G-function used in ecology to study predator-prey relationships for studying tumor-immune cell interactions in NSCLC, and 2) That a simple area under the curve (AUC) metric derived from the G-function representing tumor cell- regulatory T cell interaction correlates with poor outcome for patients, independently of clinical variables that doctors currently use! (such as age, smoking history, number of positive lymph nodes, size of tumor etc.)
Since the G-function is a unique signature of immune infiltration, my hope is that doctors will now incorporate our fast and easy-to-use algorithm in clinical practice!